Finegrained DNA research in the near future might very well contradict some of the information I have presented on the previous pages of this website. For whatever reason it may turn out that some ethnic groups passed on their genetic legacy disproportionate to their estimated share according to either historical demography or their cultural/linguistic significance. Perhaps the Cape Verdean genepool might even also include a substantial contribution from ethnic groups outside of the expected Upper Guinea area. Then again ongoing DNA research can also clarify, corroborate or even confirm the African ethnic origins for Cape Verdeans, as detailed elsewhere on this website.
There are several valid objections you might have about the presumed accuracy of personal DNA testing.1 Also the reliability of any type of scientific analysis into the genetic origins of populations should naturally not be taken for granted.2 Obviously DNA based research is still in full development. However that doesn’t imply you cannot already derive very valuable insights! Especially in the last ten years or so many meaningful improvements have been accomplished. And on condition of critical assessment and correct interpretation several highly indicative findings can already be identified.
In fact quite a few DNA studies have already been performed regarding Cape Verdean genetics. Despite its small size Cape Verde is often used by researchers as an illustrative case study with wider implications. In particular for the Atlantic Afro-Diaspora. On the following pages I will discuss different types of DNA studies. Focusing on haplogroups as well as autosomal DNA (genomewide). I will also provide a summary of my own research findings based on personal DNA test results on 23andme and Ancestry. Again all of this is naturally a work in progress and at times only providing a sketchy outlook. But sofar and all combined this DNA evidence does seem to be in line with what I discussed on the previous pages of this website.
- Haplogroup studies
- Autosomal studies (2010-2020)
- Autosomal studies (2021-2024)
- 23andme surveys
- AncestryDNA surveys
***

***
To start off this section a very early pioneering effort (1957!) to find traces of specific ethnic/regional African origins based on the bloodgroups of Cape Verdeans. This research was used by the oftquoted Cape Verdean historian Antonio Carreira to claim that the Wolof, Mandinga and Fula would be among the main ethnic elements for Cape Verdeans. However, as acknowledged by himself, there was a severe lack of samples. Something which has been true for a long time since. Fortunately in the last 10 years or so this has been changing rapidly. New research using up-to-date methodologies and based on representative reference panels is bound to deliver more insightful and also more solid outcomes.
“Em 1957, Almerindo Lessa e Jacques Ruffié publicaram, em edição da Junta de Investigações do Ultramar, um volume contendo dois trabalhos, intitulados Seroantropologia das Ilhas de Cabo Verde e Mesa Redonda sobre o Homem Cabo-Verdiano.” Source____________________
Table 1 (click to enlarge)
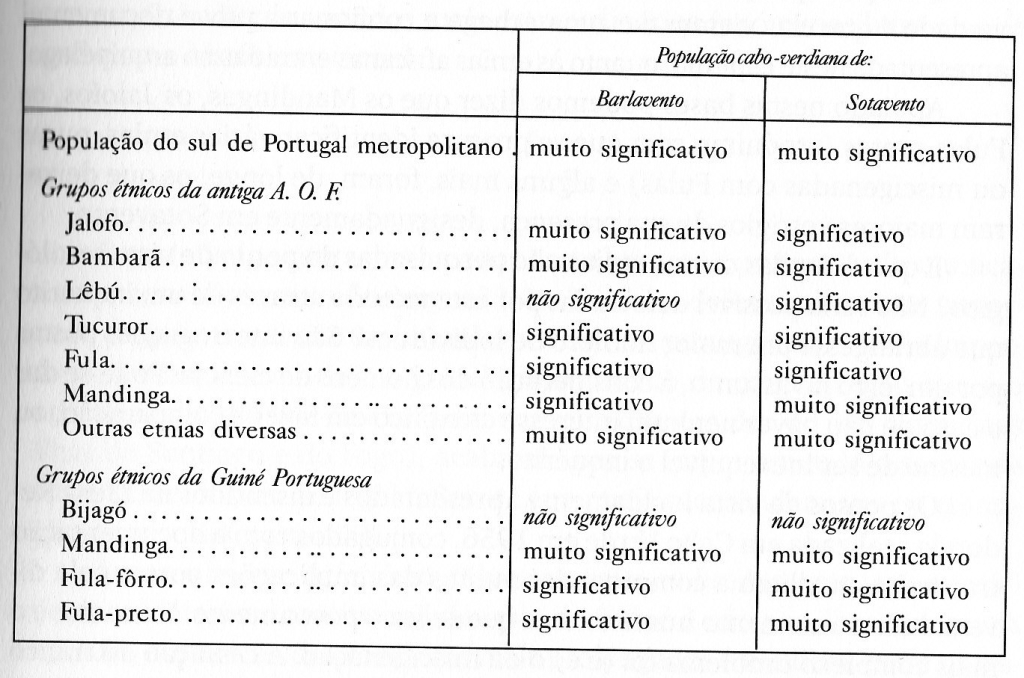
A distinction is being made between Cape Verdeans from the northern islands: Barlavento and the southern islands: Sotavento. Both of these groups show meaningful correlation with both Portuguese (southern) samples as well as most of the listed Upper Guinean samples. Intriguingly Barlavento shows higher similarity with the Wolof (Jalofo) and Bambara samples. While Sotavento shows higher similarity with Fula samples. The mainland African samples are from former French colonies in West Africa such as Senegal and Guinea Conakry (AOF = África Ocidental Francesa) as well as from Guiné Bissau. The detected relationship/correlation (based on similarity in blood group frequencies) is classified as either significant (significativo) or very significant (muito significativo). In two cases (Lebou from Senegal and Bijago from Guiné Bissau) no meaningful correlation was found. However again this might also be due to lacking sample size or other methodology issues.
___________________________________________________________________________
Notes
1) Due to source snobbery personal DNA testing results are not always fully appreciated for their informational value. I myself have never taken this overly dismissive stance. Preferring to judge each case on its own merits. From my observations especially 23andme and AncestryDNA are well-equipped to deliver results in line with the known or historically plausible backgrounds of Cape Verdeans as well as other Atlantic Afro-descendants. Of course correct interpretation and knowing how to really “read the data” remains a crucial requirement. For a more detailed discussion on the inherent restrictions of personal DNA test results see these pages:
2) The DNA studies which I will be discussing on the following pages can certainly serve as helpful tools to clarify the African ethnic origins of Cape Verdeans. However as always you do need to keep in mind inherent limitations. Don’t take everything at face value! Whenever possible countercheck their references and findings with whichever knowledge you happen to have on the topic.
Whenever I read genetic studies I always pay close attention to how the authors attempt to contextualize or explain their main research findings. As I find that historical aspects often tend to be oversimplified or even misrepresented. Given that the authors of genetic studies are usually mathematically trained naturally they tend to be focused rather on the technical aspects of whatever it is they are researching. However genetics is embedded in the social sciences as well. Especially the history of population migrations.
For those not accustomed to reading DNA studies they might seem daunting at first. Still I think that if you wish to see your personal DNA test results being placed in a wider context DNA studies can be very valuable. This is besides any general interest of course. See link below for some tips on how to “read” DNA studies.